Precise 3D modeling of the mitral valve has the potential to improve our understanding of valve morphology, particularly in the setting of mitral regurgitation (MR). Toward this goal, we have developed a user-initialized algorithm for reconstructing valve geometry from transesophageal 3D echocardiographic (3DE) image data.
Image analysis of the mitral valve at mid systole has two stages:
- user-initialized level sets segmentation
- 3D deformable modeling with continuous medial representation (cm-rep).
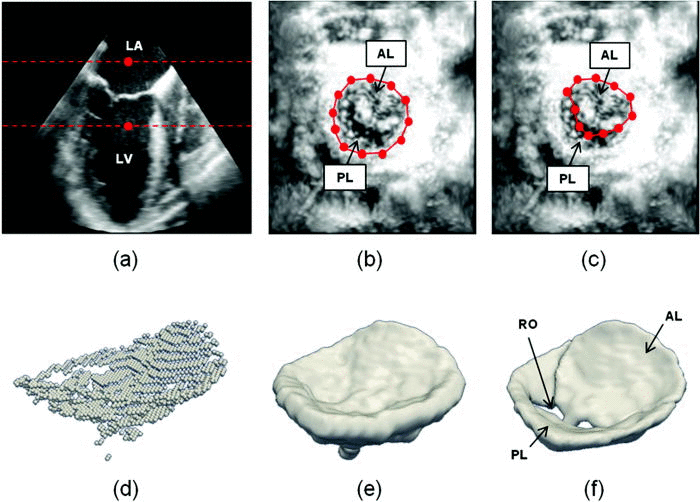
Semi-automated segmentation begins with user identification of valve location in 2D projection images generated from 3D US data. The mitral leaflets are then automatically segmented in 3D using the level set method.
- (a) The user initializes two points in a long-axis cross-section of the 3DE image volume, identifying an ROI (red) containing the valve along the axial dimension. (b) The user initializes a series of annular points in an enhanced projection image depicting the valve from an atrial perspective. (c) The user shifts posterior annular points into the coaptation zone, forming an outline of the anterior leaflet in the enhanced projection image. (d) A 3D point cloud delineating the valve is automatically generated. (e) The 3D point cloud is morphologically dilated with a spherical structuring element to obtain an ROI containing the valve. (f) A final segmentation of the valve is obtained by thresholding and active contour evolution. (LA=left atrium, LV=left ventricle, AL=anterior leaflet, PL=posterior leaflet, RO=regurgitant orifice).
Second, a bileaflet deformable medial model is fitted to the mitral leaflet segmentation by Bayesian optimization. The resulting cm-rep provides a visual reconstruction of the mitral valve, from which localized measurements of valve morphology are automatically derived.
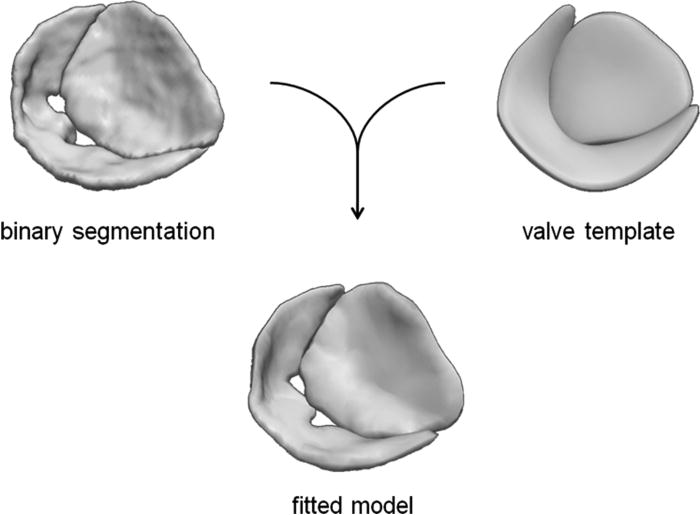
A medial representation of the valve is obtained by fitting a cm-rep template to a binary segmentation of the mitral leaflets.
Features such as anatomic regurgitant orifice area can be both visually observed and quantified from image-derived cm-reps of the mitral leaflets.
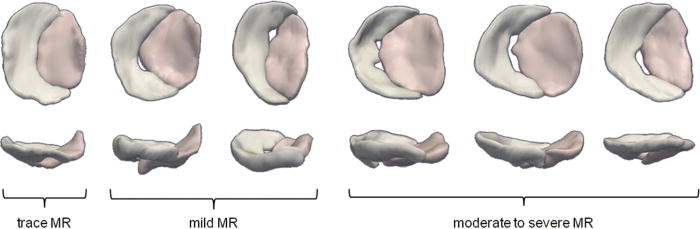
Automated reconstructions of the valves of six patients with various degrees of mitral regurgitation severity as shown from two viewpoints (atrial and lateral perspectives).