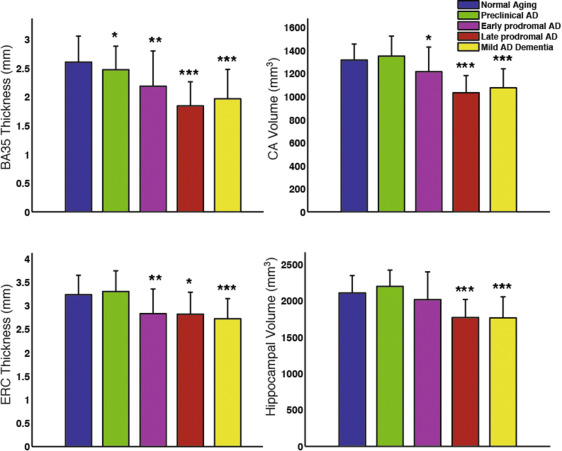
Group comparison of select MTL subregional measurements adjusted for age and ICV. Statistical significance is based on comparison with the “normal aging” group (amyloid-negative ADNI controls). Error bars reflect 1 standard deviation. *: p<0.05, **: p<0.01, ***:p<0.001.
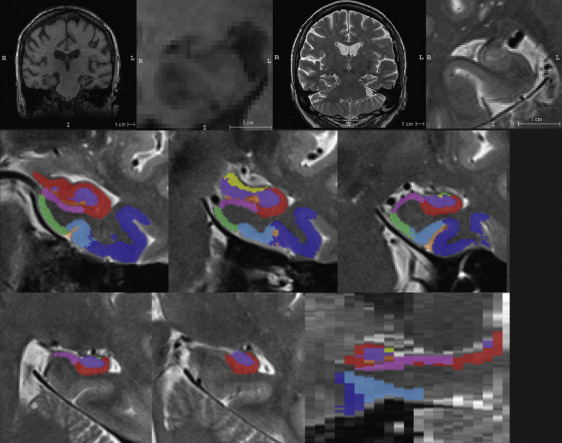
Top row from left to right: ADNI T1-MRI, T1-MRI zoomed in to hippocampus in coronal view, ADNI T2-MRI, zoomed in to hippocampus in oblique coronal view. Middle and bottom row show an example of automatic segmentation of MTL subregions using ASHS.
Recently published paper by Wolk et al. in Neurobiology of Aging examines the involvement of hippocampal subfields and medial temporal lobe (MTL) cortical subregions in different stages of Alzheimer’s disease (AD). The study used high-resolution T2-weighted MRI scans of the hippocampal region from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) to perform pseudo-longitudinal analysis of the continuum from preclinical to mild AD dementia, with the groups were further divided according to amyloid status based on PET. The patterns of atrophy in the MTL subregions largely recapitulated Braak staging of neurofibrillary tangles (NFT) within the MTL. Particularly, Brodmann Area 35 (the first region associated with NFT deposition), was the only region to discriminate preclinical AD from amyloid negative CN adults (“normal aging”). These findings suggest that subregional measures derived automatically from high-resolution MRI scans with ASHS may be a valuable biomarker for therapeutic trials in AD and related neurodegenerative disorders.