Comprehensive visual and quantitative analysis of in vivo human mitral valve morphology is central to the diagnosis and surgical treatment of mitral valve disease. Real-time 3D transesophageal echocardiography (3D TEE) is a practical, highly informative imaging modality for examining the mitral valve in a clinical setting. To facilitate visual and quantitative 3D TEE image analysis, we describe a fully automated method for segmenting the mitral leaflets in 3D TEE image data. The algorithm integrates complementary probabilistic segmentation and shape modeling techniques:
- multi-atlas joint label fusion, and
- deformable modeling with continuous medial representation
to automatically generate 3D geometric models of the mitral leaflets from 3D TEE image data. These models are unique in that they establish a shape-based coordinate system on the valves of different subjects and represent the leaflets volumetrically, as structures with locally varying thickness. Without any user interaction, the method accurately captures patient-specific leaflet geometry at both systole and diastole in 3D TEE data acquired from a mixed population of subjects with normal valve morphology and mitral valve disease.
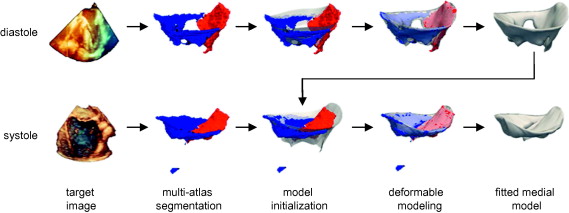
Automatic segmentation of the mitral leaflets at diastole (top row) and systole (bottom row) for a given patient. First, a probabilistic segmentation is generated by multi-atlas label fusion (red = anterior leaflet, blue = posterior leaflet). Then the cm-rep template (translucent) is initialized to the multi-atlas segmentation and the template is deformed to obtain a medial model of the mitral leaflets. The fitted diastolic model is used to initialize model fitting of the same subject’s valve at systole.